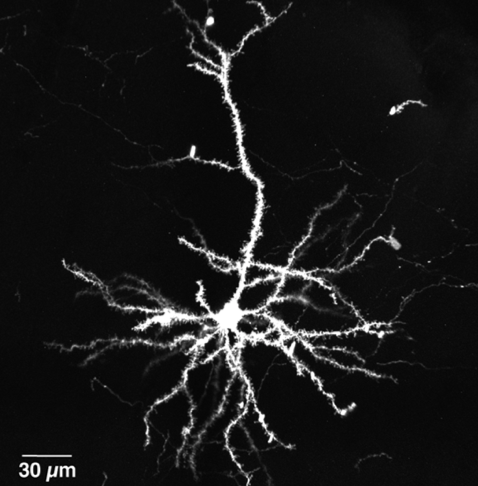
A challenging aspect of neuroscience revolves around mapping the synaptic connections within neural circuits (connectomics) over scales spanning several orders of magnitude (nanometers to meters). Despite significant improvements in serial section electron microscopy (SSEM) technologies, several major roadblocks have impaired its general applicability to mammalian neural circuits. In the present study, we introduce a new approach that circumvents some of these roadblocks by adapting a genetically-encoded ascorbate peroxidase (APEX2) as a fusion protein to a membrane-targeted fluorescent reporter (CAAX-Venus), and introduce it in single pyramidal neurons in vivo using extremely sparse in utero cortical electroporation. This approach allows us to perform Correlated Light-SSEM (CoLSSEM), a variant of Correlated Light-EM (CLEM), on individual neurons, reconstructing their dendritic and axonal arborization in a targeted way via combination of high-resolution confocal microscopy, and subsequent imaging of its ultrastructural features and synaptic connections with ATUM-SEM (automated tape-collecting ultramicrotome - scanning electron microscopy) technology. Our method significantly will improve the feasibility of large-scale reconstructions of neurons within a circuit, and permits the description of some ultrastructural features of identified neurons with their functional and/or structural connectivity, one of the main goal of connectomics.